


|
|
|
Target-miRNA |
The program Target-miRna is developed for search for microRNA (miRNA) sites in genomic sequences. miRNAs promote mRNA cleavage at almost perfect complementarity to its site. In case of less complementarity, miRNAs inhibit mRNA translation. Our program Target-miRna searches a given target sequence for microRNA sites, basing on calculation of the interaction energy between miRNA and its site. Therefore Target-miRna can be used for search of both site types.
Target-miRna scans a target sequence and calculates the energy of complementary interaction between miRNA and possible site i as follows:
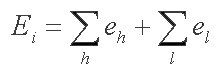
where eh is helix energy and el is loop energy if any.
The energy parameters of complementary interactions and loops are taken from Turner's table. To skip suboptimal miRNA-site pairing we minimize the interaction energy by a dynamic algorithm which is based on Nussinov and Jackobson and Zuker papers. The user sets an energy threshold, and Target-miRna outputs all the candidate sites, which energy of miRNA-site interaction is lower (i.e., more stable) than it.
Target-miRna supports two different search modes. In the first mode the user inputs a single miRNA sequence by himself.
In the second mode the user specifies the organism and our program searches for the sites for all miRNAs known
for this organism, using built-in miRNA library. Currently the library contains the miRNAs of the following organisms:
cel (Caenorhabditis elegans)
hsa (Homo sapiens)
dme (Drosophila melanogaster)
mmu (Mus musculus)
ath (Arabidopsis thaliana)
rno (Rattus norvegicus)
oza (Oryza sativa)
ebv (Epstein Barr)
gga (Gallus gallus)
dps (Drosophila pseudoobscura)
dre (Danio rerio)
xla (Xenopus laevis)
zma (Zea mays)
sbi (Sorghum bicolor)
ame (Apis mellifera)
aga (Anopheles gambiae)
cfa (Canis familiaris)