


|
|
|
LCRep |
Program for mapping low complexity regions in nucleotide sequences.
Search for the low complexity regions is performed with using Shannon's information measure. Shannon's information is defiened as follows:
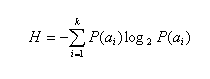
where:
{a1, ..., ak} is the alphabet of the size k, and P(ai) is a
fractional composition of ai
The search is carried out as follows. For each position i of the sequence S calculation of the Shannon's information H(i, l) is performed in the window of size l within the range [lbegin, lend]. If H(i, l) turns out below prespecified threshold Hthr(l) then fragment [i, i+l] is declared low complex. Intersection of all such fragments at the end of calculation gives a map of low complexity regions of the sequence S.
>c20 Masked regions: p1: 90 p2: 115 l: 26 chain(+) [Low Complexity Region] p1: 220 p2: 240 l: 23 chain(+) [Low Complexity Region] ....
>c20 GCCAAGAAGATATGTAGCATTAAGGTTTAGAATACAGGCTTTGAAGTCAAACAGACCAGAGTTAACAACCTCATTTTGTT TTTATTTTCNNNNNNNNNNNNNNNNNNNNNNNNNNCTTTAAGTTCTAGGGTACATGTGCACAACGTGCAGGTTTGTTACA TATGTATACATGTGCCATGTTGGTGTGCTGCACCCATTAACTGGACATTTACATTAGGTNNNNNNNNNNNNNNNNNNNNN CCCTCCTCCCCTTACCCCACAACAGGCCCCGGTGTGTGATGTTCCCCTTCCTGTGTCCAAGTGTTCTCATTGTTCAGTTC ....
>c20 gccaagaagatatgtagcattaaggtttagaatacaggctttgaagtcaaacagaccagagttaacaacctcattttgtt tttattttcTTTTTTAAAATTTTTTTAAAATTATActttaagttctagggtacatgtgcacaacgtgcaggtttgttaca tatgtatacatgtgccatgttggtgtgctgcacccattaactggacatttacattaggtAAAAAAAAAAAAAAAAAAAAA ccctcctccccttaccccacaacaggccccggtgtgtgatgttccccttcctgtgtccaagtgttctcattgttcagttc ....