


|
|
|
GeneCorr |
The program calculates correlation coefficients between the gene expression profiles.
Program is provided with viewer.
The expression data for the set of genes is represented as a table, consisting of rows (usually corresponding to genes) and columns (or fields, usually corresponding to samples/tissues/experiments). Each row corresponds to expression measurements for the gene. Columns correspond to experiments/samples/tissues. However, this table may include not only expression data, but also other information related to genes, for example gene names, classifiers, etc. Therefore we will call the table columns as 'fields' in general case. In general, columns of the table could be of four basic types:
| IVALUE | signed integer value; | |
| FVALUE | floating point value; | |
| WORD | text without spaces inside (single word); | |
| STRING | text with spaces inside allowed. |
Basic input file format should be as follows:
; May contain comment starting from the semicolon in any line of the file NAME<tab>WORD GENEID<tab>IVALUE TISSUECANCER0<tab>FVALUE TISSUECANCER1<tab>FVALUE TISSUENORMAL0<tab>FVALUE TISSUENORMAL1<tab>FVALUE TISSUENORMAL2<tab>FVALUE #GROUP<tab>Cancer tissues TISSUECANCER0 TISSUECANCER1 #ENDGROUP #GROUP<tab>Arbitrary group TISSUECANCER1 TISSUECANCER2 TISSUENORMAL0 TISSUENORMAL1 #ENDGROUP END DATA GENE04675<tab>402<tab>6.00<tab>5.60<tab>5.97<tab>6.00<tab>6.00 GENE46890<tab>794<tab>2.77<tab>3.22<tab>5.65<tab>5.68<tab>5.68 GENE23794<tab>404<tab>5.97<tab>5.97<tab>6.00<tab>5.60<tab>5.97
In this example <tab> implies 'Tab' character symbol.
First lines (up to the "DATA" line) contain data format description. In this part of the file each line describes field description: field name and field basic type.
After the "DATA" line - data on each gene are represented. Each line correspond single cards. Field data are separated by 'tab' symbol. Double 'tab' is interpreted as missed data.
It is assumed in SetTag program that the expression data in the file are normalized and the expression levels of genes in experiments are comparable.
MolQuest version of the SelTag program can also operates with other types of files, namely, selection files. These files contain information about some selected genes or samples from the large data file in SelTag format. The selection file contain: the data file name from which selection was obtained; type of selection data (genes of samples), list of selected objects (their indices in the large data file). The selection files are in the XML format. Two examples are below.
Selection for some genes.
<?xml version="1.0" encoding="ISO-8859-5"?> <SELECTION> <HEADER name="cc_Selection5"> <DATA source="c:/data/cc.txt"/> <COMMENT><![CDATA["$F1 == "GEN14263" | $F12 >= 300"]]></COMMENT> </HEADER> <ELEMENTS type="GENES" count="9"> <![CDATA[0;1;2;10;14;15;17;26;30]]> </ELEMENTS> </SELECTION>
Selection for some fields (samples).
<?xml version="1.0" encoding="ISO-8859-5"?> <SELECTION> <HEADER name="notterman2001_set1"> <DATA source="c:/data/notterman2001_set1.txt"/> <COMMENT><![CDATA["From cc.txt data file."]]></COMMENT> </HEADER> <ELEMENTS type="FIELDS" count="10"> <![CDATA[0;1;2;3;5;6;7;18;19;30]]> </ELEMENTS> </SELECTION>
Selection files may be selected during the SelTag execution and also used by SelTag for calculation and/or visualization. Note, each selection file is linked to large data file by its name. Selection data cannot be applied to another data file.
User should define two lists of genes, program will calculate correlation coefficients between gene expression profiles from different lists. User can also set the threshold for correlation value to select most correlated pairs.
User should provide list of fields to calculate correlation.
Three types of correlation are possible:
Pearson's r - Pearson's correlation coefficient.
The Pearson product moment correlation coefficient between expression profiles i and j is calculated as follows:
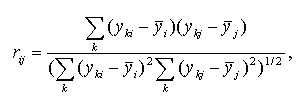
where yki is the expression level of gene i in the experiment k;
![]() is the mean
expression level of the gene i. Positive correlation implies that the
expression levels of genes i,j are related positively, the higher expression
of gene i, the higher expression of gene j. Negative correlation
means that the expression levels of genes i,j are related negatively, the
higher expression of gene i, the lower expression of gene j.
If the rij is close to zero, two expression profiles are unrelated.
is the mean
expression level of the gene i. Positive correlation implies that the
expression levels of genes i,j are related positively, the higher expression
of gene i, the higher expression of gene j. Negative correlation
means that the expression levels of genes i,j are related negatively, the
higher expression of gene i, the lower expression of gene j.
If the rij is close to zero, two expression profiles are unrelated.
Spearman r - Spearman's correlation coefficient.
This correlation coefficient is computed for ranks. Let Rki is
the rank of the expression level in the experiment k of gene i
(relatively to other experiments), Rkj is the rank of the
expression level in the experiment k of gene j.
Then Spearman's correlation coefficient is calculated by the formula
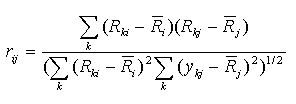
Kendall's t - Kendall's tau correlation coefficient.
To calculate Kendall's t K for data points
(yki; ykj) 2K(K - 1) pairs considered (without self-pairing, the points
in either order count as one pair).
Pairs in which yki > ymi and
ykj > ymj or
yki < ymi and ykj < ymj
are called concordant pairs (agreement between ranks), pairs with rank disagreement are called discordant pairs.
In general, t is calculated as
t = ([number of concordant] - [number of discordant]) / total number of pairs
Correlation coefficients (Spearman rank correlation) between gene expression profiles: List1\List2 GEN30482 GEN03437 GEN30823 GEN01998 0.5657 0.4885 0.4939 GEN03687 0.7642 0.7814 0.7617 GEN24649 0.5858 0.5624 0.6399 GEN09108 0.1657 0.0949 -0.1042 GEN09514 0.4313 0.3925 0.2861 GEN02303 0.5876 0.5993 0.4568 List of gene pairs with the absolute value of the correlation coefficients above threshold (0.7722) GEN03687 GEN03437 : 0.7814 GEN02374 GEN03437 : 0.7941 GEN02374 GEN30823 : 0.8520
First line is the header. It contains the type of the calculated correlation in parentheses. Second line is the list if gene identifiers from the List1, separated by tabulation. Next lines list data for genes for List2 separated by tabulation.